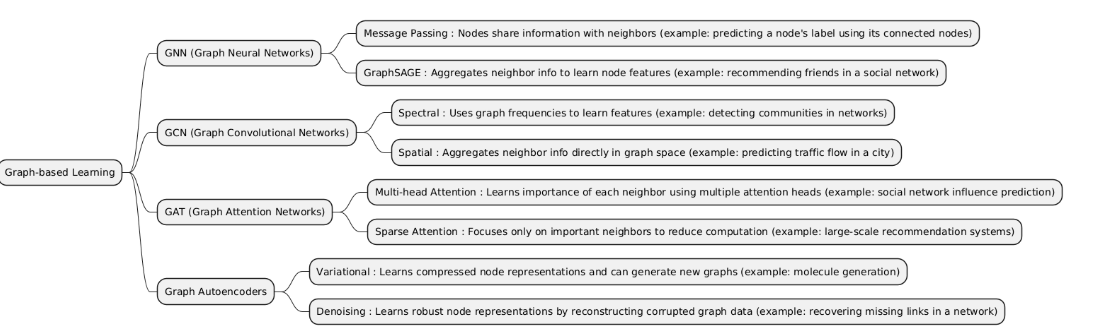
Graph-based Learning is a type of machine learning that focuses on data represented as graphs, where nodes represent entities and edges represent relationships. It is used to capture the structure and connections in data, enabling tasks like node classification, link prediction, and network analysis.
| Type | What it is | When it is used | When it is preferred over other types | When it is not recommended | Examples of projects that is better use it incide him |
|---|---|---|---|---|---|
| GNNs | Graph Neural Networks (GNNs) are deep learning models designed to operate on graph-structured data, learning node, edge, or graph-level representations by aggregating information from neighbors. | Used for tasks involving relational data, such as social networks, molecules, knowledge graphs, or transportation networks. |
• Better than GCN/GAT/Graph Autoencoders when you want a general-purpose graph model without a specific convolution or attention mechanism. • Ideal for initial experiments on graph data to understand node and graph relationships. |
• When you need specialized operations, like convolution (GCN) or attention (GAT) — those may perform better. • When the graph is small or simple — traditional ML methods may suffice. • For non-graph-structured data — CNNs, RNNs, or Transformers are better. |
• Node classification in a social network (predict user interests). • Link prediction in recommendation systems (predict friendships or connections). • Molecule property prediction in drug discovery. • Traffic flow prediction using road network graphs. |
| GCNs | Graph Convolutional Networks (GCNs) are a type of Graph Neural Network that applies convolution-like operations on graph nodes, aggregating features from each node’s neighbors to learn node or graph-level embeddings. | Used for tasks on graph-structured data, especially when local neighborhood information is important, such as node classification, link prediction, or graph classification. |
• Better than general GNNs when you want explicit convolution operations for better local feature aggregation. • Better than GATs when attention mechanisms are unnecessary and simpler aggregation suffices. • Better than Graph Autoencoders when discriminative tasks (classification or prediction) are the goal rather than reconstruction. |
• For graphs where different neighbor importance matters — GAT may perform better. • When the task is unsupervised graph representation learning — Graph Autoencoders are more suitable. • For non-graph data — CNNs, RNNs, or Transformers are better. |
• Node classification — predicting types of users in a social network. • Link prediction — recommending new connections in a network. • Graph classification — predicting chemical properties of molecules. • Fraud detection — detecting suspicious transactions in financial networks. |
| GATs | Graph Attention Networks (GATs) are a type of Graph Neural Network that use attention mechanisms to assign different weights to neighboring nodes when aggregating features, allowing the model to focus on the most relevant neighbors. | Used for graph-structured tasks where neighbor importance varies, such as node classification, link prediction, and graph-level prediction. |
• Better than GNNs/GCNs when not all neighbors contribute equally, and attention can improve performance. • Better than Graph Autoencoders for discriminative tasks rather than unsupervised reconstruction. • Preferred over GCNs when graph connectivity is irregular or highly variable. |
• When graph is small or uniform, GCNs may suffice. • When computational resources are limited, as attention increases complexity. • For unsupervised embedding or reconstruction tasks — Graph Autoencoders are better. |
• Node classification in social networks with variable connection importance. • Fraud detection in financial networks where certain connections matter more. • Molecule property prediction focusing on key atomic interactions. • Recommendation systems weighting important user-item links. |
| Graph Autoencoders | Graph Autoencoders are a type of Graph Neural Network designed for unsupervised learning on graph-structured data. They encode nodes into a latent space and then reconstruct the graph structure (adjacency matrix) or node features. | Used for graph embedding, link prediction, and anomaly detection on graphs. |
• Better than GNNs/GCNs/GATs when the task is unsupervised representation learning rather than classification or prediction. • Preferred over GATs/GCNs when you want graph reconstruction or low-dimensional embeddings for downstream tasks. |
• For discriminative tasks like node classification — GCNs or GATs are better. • When computational efficiency is critical — simpler GCNs may suffice. • For non-graph data — CNNs, RNNs, or Transformers are more appropriate. |
• Link prediction — predicting missing edges in social networks. • Graph embedding — compressing large graphs for visualization or clustering. • Anomaly detection — identifying unusual nodes or edges in a network. • Recommendation systems — embedding users/items to predict interactions. |
import networkx as nx
import numpy as np
# --- 1. Create a simple graph ---
G = nx.Graph()
G.add_edges_from([
(0, 1), (0, 2), (1, 2), (1, 3),
(2, 3), (3, 4)
])
# --- 2. Initialize node features ---
features = {
0: np.array([1.0, 0.0]),
1: np.array([0.0, 1.0]),
2: np.array([1.0, 1.0]),
3: np.array([0.5, 0.5]),
4: np.array([0.0, 0.0])
}
# --- 3. GraphSAGE aggregation function ---
def graphsage_aggregate(node, features, G):
neighbors = list(G.neighbors(node))
if len(neighbors) == 0:
return features[node]
neighbor_feats = np.array([features[n] for n in neighbors])
agg = neighbor_feats.mean(axis=0) # mean aggregation
# Combine node's own feature with aggregated neighbor features
new_feature = np.tanh(features[node] + agg) # simple non-linearity
return new_feature
# --- 4. Update features for all nodes ---
new_features = {}
for node in G.nodes():
new_features[node] = graphsage_aggregate(node, features, G)
# --- 5. Show new features ---
for node, feat in new_features.items():
print(f"Node {node} new feature: {feat}")
import networkx as nx
import numpy as np
# --- 1. Create a simple graph ---
G = nx.Graph()
G.add_edges_from([
(0, 1), (0, 2), (1, 2), (1, 3),
(2, 3), (3, 4)
])
# --- 2. Initialize node features ---
features = {
0: np.array([1.0, 0.0]),
1: np.array([0.0, 1.0]),
2: np.array([1.0, 1.0]),
3: np.array([0.5, 0.5]),
4: np.array([0.0, 0.0])
}
# --- 3. Simple attention function ---
def attention(query, key):
# simple dot-product attention
score = np.dot(query, key)
return np.exp(score)
# --- 4. Multi-head GAT aggregation ---
def multihead_gat(node, features, G, heads=2):
neighbors = list(G.neighbors(node))
if len(neighbors) == 0:
return features[node]
head_outputs = []
for h in range(heads):
# apply random linear transformation per head
W = np.random.randn(2, 2) * 0.5
node_feat = np.dot(features[node], W)
neighbor_feats = np.array([np.dot(features[n], W) for n in neighbors])
# compute attention scores
scores = np.array([attention(node_feat, nf) for nf in neighbor_feats])
scores = scores / scores.sum() # normalize
# aggregate neighbor features weighted by attention
agg = (scores[:, None] * neighbor_feats).sum(axis=0)
head_outputs.append(np.tanh(node_feat + agg)) # combine with own feature
# Concatenate outputs from all heads
return np.concatenate(head_outputs)
# --- 5. Update features for all nodes ---
new_features = {}
for node in G.nodes():
new_features[node] = multihead_gat(node, features, G, heads=2)
# --- 6. Show new features ---
for node, feat in new_features.items():
print(f"Node {node} new feature: {feat}")